Anatomical Landmark Detection From X-Ray Images Using Convolutional Neural Networks
Abstract
In this thesis, we propose an approach that robustly detects three-dimensional (3D) anatomical landmarks in two-dimensional (2D) radiography projections (X-rays). 2D X-ray images are widely used since they provide a fast and accurate assessment of bony anatomies.Correctly identifying specific 3D landmarks in the images is crucial for clinical applica- tions, e.g., planning of total hip replacement (THR).
Compared to 3D medical imaging such as computed tomography (CT), clinical evaluations in radiography projections are limited to the 2D plane. During X-ray acquisition, occurring pose variations result in perspective distortions in the image. Moreover, anatomical structures are usually projected such that the order of appearance along the X-ray beam is ambiguous. Therefore, accurately detecting 3D points of interest based solely on the 2D projections is a complex task.
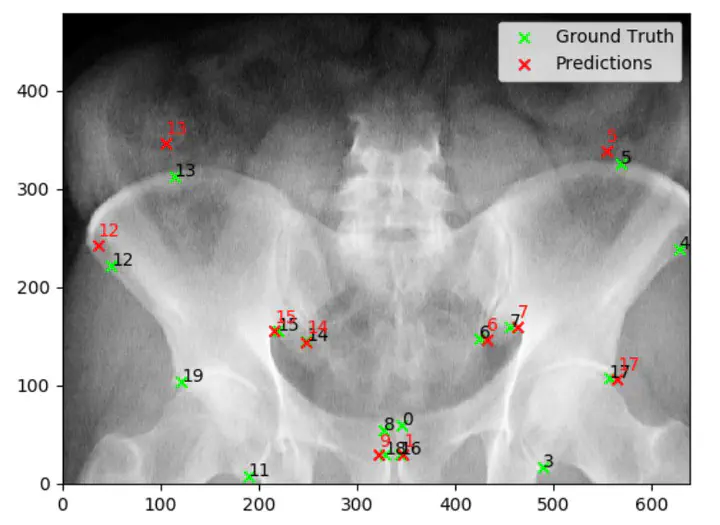
To compensate for the aforementioned distortions, we incorporate a synthetic data generation framework that defines 3D landmarks on the anatomies of interest in CT images. The landmarks and CT intensity information are projected in the 2D space and used to train a convolutional neural network (CNN) with a heatmap regression approach. Using synthetic heatmaps as ground truth for landmark locations drives the CNN to produce heatmap-like activations centered at the 2D positions of the underlying 3D anatomical landmark. We further apply regularization via articulated statistical shape and intensity models (A-SSIMs) to obtain an estimate of the third-dimensional component of the detected landmark (i.e. landmark back-projection).
The proposed methods for CNN landmark detection and back-projection are evaluated based on artificial digitally reconstructed radiographs (DRRs) from pelvic CTs as well as clinical X-ray images of the hip joint. The results indicate that CNNs only trained on synthetic data generalize to unseen clinical X-ray images for 2D landmark prediction. We further believe that the back-projected landmarks obtained via A-SSIMs can serve as initialization points for intensity-based 2D/3D reconstruction and can be useful for gaining an insight into the 3D shape of the depicted anatomy.